9 Introduction
9.1 Population and Sample
A population refers to the entire group of individuals or instances about whom we hope to learn. It encompasses all possible subjects or observations that meet a set of criteria. The population is the complete set of items that interest the researcher, and it can be finite (e.g. the students in a particular school) or infinite (e.g. the number of times a die can be rolled). A population size is given by the number of distinct elements and it includes every individual or observation of interest.
A sample is a subset of the population that is used to represent the population. Since studying an entire population is often impractical due to constraints like time, cost, and accessibility, samples provide a manageable and efficient way to gather data and make inferences about the population. It is important that the sample is representative of the population of interest to allow for valid inferences. It is always important to distinguish between a random sample, e.g. a random group of students in 5th year from a school to make inference about the students at the 5th year of such school, and a convenience sample, e.g. a class of 5th year students who are easily accessible to the researcher, but that can be not representative of all the 5th year students in the school.
| Aspect | Population | Sample |
|---|---|---|
| Definition | Entire group of interest | Subset of the population |
| Size | Large, potentially infinite | Small, manageable |
| Data Collection | Often impractical to study directly | Practical and feasible |
| Purpose | To understand the whole group | To make inferences about the population |
Any finite sample is a collection of realization of a finite number of random variables. For example, consider a set of random vector
In general, we distinguish between finite or non-finite populations. In the case of a finite population with
- With reimmission of
- Without readmission of
9.2 Estimators
Let’s consider a statistical model depending on some unknown parameter
Since the estimator is a random variable itself, one can define some metrics to compare different estimators of the same parameter. Firstly, let’s consider the bias, the distance between the average of the collection of estimates and the single parameter being estimated, i.e.
9.2.1 Properties
As for some desirable property of an estimator one have
- Consistency: An estimator
Example 9.1 Let’s consider an IID normally distributed sample, i.e.
A. Sample mean: The natural estimator for
B. Biased but consistent estimator. Suppose we estimate
Even if
C. Inconsistent estimator: Let’s define as estimator for
Example: consistency of the sample mean
library(dplyr)
set.seed(1)
# =======================================
# Inputs
# =======================================
# True parameters
par <- c(mu = 2, sigma = 2)
# True moments (normal)
moments <- c(par[1], par[2])
# Observations for each sample
n <- c(5, 10, 100, 200, 300, 400, 500)
# Number of samples
n.sample <- 1000
# Confidence interval
alpha <- 0.005
# =======================================
# Consistency function
example_consistency <- function(n = 100, n.sample = 1000, par, moments, alpha = 0.005){
estimates <- list()
for(i in 1:n.sample){
x_n <- rnorm(n, par[1], par[2])
# Case A
mu_A <- sum(x_n) / n
# Case B
mu_B <- mu_A / (n+1) * n
# Case C
mu_C <- x_n[1]
estimates[[i]] <- dplyr::tibble(i = i, n = n,
A = mu_A,
B = mu_B,
C = mu_C,
dw = qnorm(alpha, moments[1], moments[2]/sqrt(n)),
up = qnorm(1-alpha, moments[1], moments[2]/sqrt(n)))
}
dplyr::bind_rows(estimates) %>%
group_by(n) %>%
mutate(e_A = mean(A), q_dw_A = quantile(A, alpha), q_up_A = quantile(A, 1 - alpha),
e_B = mean(B), q_dw_B = quantile(B, alpha), q_up_B = quantile(B, 1 - alpha),
e_C = mean(C), q_dw_C = quantile(C, alpha), q_up_C = quantile(C, 1 - alpha))
}
# =======================================
# Generate data
data <- purrr::map_df(n, ~example_consistency(.x, n.sample, par, moments, alpha)) Code
library(ggplot2)
fig_A <- ggplot(data)+
geom_point(aes(n, A))+
geom_line(aes(n, moments[1], color = "theoric"))+
geom_line(aes(n, e_A, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, dw, color = "theoric"))+
geom_line(aes(n, up, color = "theoric"))+
geom_line(aes(n, q_up_A, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, q_dw_A, color = "empiric"), linetype = "dashed")+
theme_bw()+
scale_color_manual(
values = c(theoric = "red", empiric = "purple"),
labels = c(theoric = "Theoric", empiric = "Empiric")
)+
theme(legend.position = "none")+
labs(x = "", y = "", subtitle = "A", color = "")+
scale_y_continuous(limits = range(c(data$A, data$B, data$C)))
fig_B <- ggplot(data)+
geom_point(aes(n, B))+
geom_line(aes(n, moments[1], color = "theoric"))+
geom_line(aes(n, e_B, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, dw, color = "theoric"))+
geom_line(aes(n, up, color = "theoric"))+
geom_line(aes(n, q_up_B, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, q_dw_B, color = "empiric"), linetype = "dashed")+
theme_bw()+
scale_color_manual(
values = c(theoric = "red", empiric = "purple"),
labels = c(theoric = "Theoric", empiric = "Empiric")
)+
theme(legend.position = "none")+
labs(x = "Sample size", y = "", subtitle = "B", color = "")+
scale_y_continuous(limits = range(c(data$A, data$B, data$C)))
fig_C <- ggplot(data)+
geom_point(aes(n, C))+
geom_line(aes(n, moments[1], color = "theoric"))+
geom_line(aes(n, e_C, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, dw, color = "theoric"))+
geom_line(aes(n, up, color = "theoric"))+
geom_line(aes(n, q_up_C, color = "empiric"), linetype = "dashed")+
geom_line(aes(n, q_dw_C, color = "empiric"), linetype = "dashed")+
theme_bw()+
scale_color_manual(
values = c(theoric = "red", empiric = "purple"),
labels = c(theoric = "Theoric", empiric = "Empiric")
)+
theme(legend.position = "none")+
labs(x = "", y = "", subtitle = "C", color = "")+
scale_y_continuous(limits = range(c(data$A, data$B, data$C)))
gridExtra::grid.arrange(fig_A, fig_B, fig_C, nrow = 1)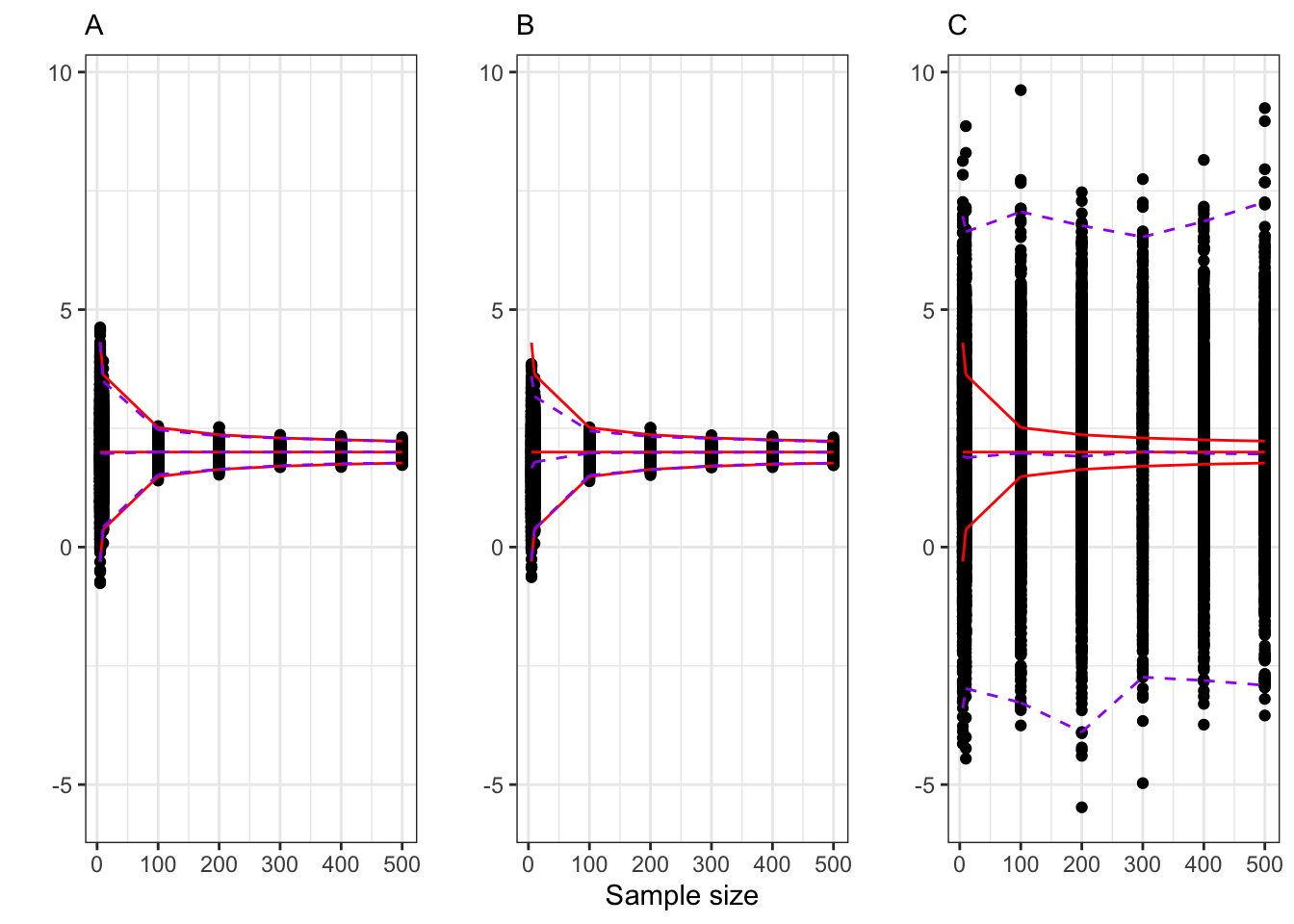
Efficiency: Among unbiased estimators, the one with minimal variance is called efficient; asymptotically efficient means it attains the Cramér–Rao lower bound (CRLB) limit.
Asymptotic normality: for some normalizing sequence
9.2.2 Sufficiency and Completeness
Theorem 9.1 (
A statistic
In other words, a statistic is sufficient if it captures all the information in the sample about
Example 9.2 If
Theorem 9.2 (
If
In other words, conditioning on a sufficient statistic cannot increase variance. So Rao–Blackwell gives a systematic way to improve an estimator: start with any unbiased estimator, then condition it on a sufficient statistic to possibly reduced the variance.
Definition 9.1 (
A statistic
Intuition: if a function of a statistic always averages out to zero no matter what the parameter is, then it must be the trivial zero function. Completeness rules out the existence of “hidden functions” of the statistic that are unbiased estimators of
Example 9.3 Continuing from Example 9.2, the statistic
Theorem 9.3 (
If
In practice, sufficiency (Theorem 9.1) ensures
Example 9.4 Continuing from Example 9.3,