27 Value at Risk test
Let’s consider a ARMA(2,2)-GARCH(1,1) model defined as:
27.1 Normal distribution
Let’s consider independent and normally distributed residuals
27.2 Gaussian Mixture distribution
Let’s consider independent and Gaussian mixture distributed residuals
27.3 Test on the number of violations
Let’s define a violation of the
and let’s define the number of violations of the conditional VaR as follows, i.e.
27.3.1 Asymptotic variance
Applying the central limit theorem (CLT) it is possible to prove that the statistic test converges in distribution to a standard normal, i.e.
where
27.3.2 Empirical variance
Instead of using the theoretical variance of
For small samples, the following relation between the two statistics should be used:
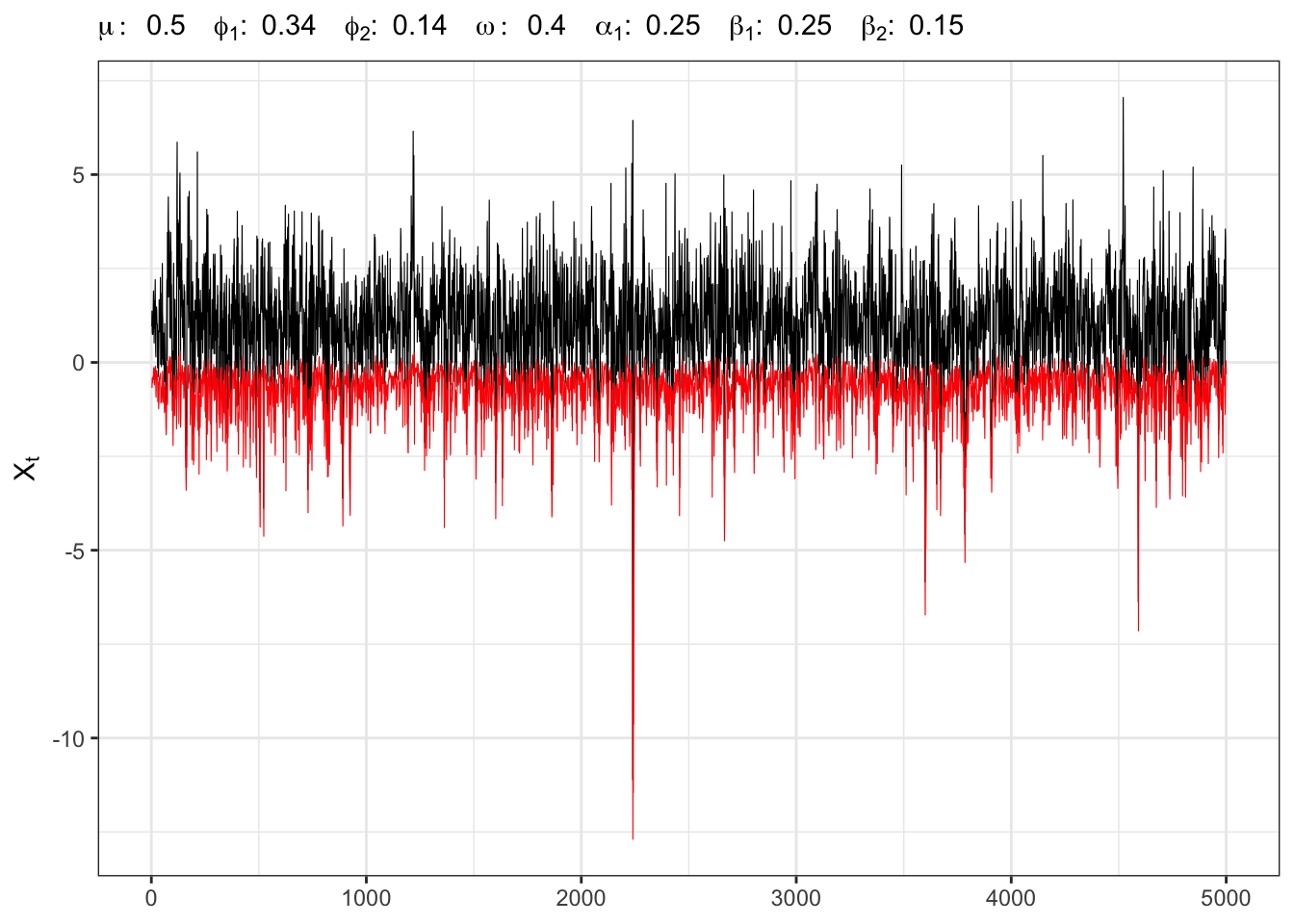
27.4 Example:
Instead of simulating exactly
AR(2)-GARCH(1,2) simulation and VaR
# ===================== Setups ======================
set.seed(1) # random seed
ci <- 0.05 # confidence level
t_bar <- 5000 # number of simulations
# parameters
parAR <- c(mu=0.5, phi1 = 0.34, phi1 = 0.14)
parGARCH <- c(omega=0.4, alpha1=0.25,
beta1=0.25, beta2=0.15)
# long-term std deviation of residuals
sigma_eps <- sqrt(parGARCH[1]/(1-sum(parGARCH[-1])))
# ================== Simulation =====================
# Initial points
Xt <- rep(parAR[1]/(1-sum(parAR[-1])), 3)
sigma <- rep(sigma_eps, 3)
# Simulated residuals
eps <- rt(t_bar, 25)
eps[1:3] = eps[1:3]*sigma_eps
# Value at Risk
q_alpha = qnorm(ci)
VaR = c(0)
for(t in 3:t_bar){
# AR component
Xt[t] <- parAR[1] + parAR[2]*Xt[t-1] + parAR[3]*Xt[t-2]
# ARCH component
sigma[t] <- parGARCH[1] + parGARCH[2]*eps[t-1]^2
# GARCH component
sigma[t] <- sigma[t] + parGARCH[3]*sigma[t-1]^2 + parGARCH[4]*sigma[t-2]^2
sigma[t] <- sqrt(sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated value at risk
VaR[t] <- Xt[t] + sigma[t]*q_alpha
# Simulated time series
Xt[t] <- Xt[t] + eps[t]
}
# ===================== Plot ======================
# GARCH(1,1) simulation
ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, VaR), size = 0.2, color = "red")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", parAR[1],
"\\;\\; \\phi_{1}:\\;", parAR[2],
"\\;\\; \\phi_{2}:\\;", parAR[3],
"\\;\\; \\omega:\\;", parGARCH[1],
"\\;\\; \\alpha_{1}:\\;", parGARCH[2],
"\\;\\; \\beta_{1}:\\;", parGARCH[3],
"\\;\\; \\beta_{2}:\\;", parGARCH[4],
"$")))VaR test Student-
# ======================================
# Violation of the VaR
vt <- ifelse(Xt < VaR, 1, 0)
vt[1:3] <- 0
# Theoric variance
v_theoric <- ci*(1-ci)
# Empiric variance
v_empiric <- (sum(vt)/t_bar)*(1 - sum(vt)/t_bar)
# Standardized number of violations
Nt <- sum((vt - ci)/sqrt(v_theoric))
# Statistic test (NV_1)
T1 <- (1/sqrt(t_bar))*Nt
# Statistic test (NV_2)
T2 <- (sqrt(v_theoric)/sqrt(v_empiric))*T1
# Rejection level
t_alpha <- qnorm(ci/2)
# =============== Kable ===============
kab <- dplyr::tibble(
n = t_bar,
alpha = paste0(format(ci*100, digits = 3), "%"),
alpha_hat = paste0(format(sum(vt)/t_bar*100, digits = 3), "%"),
t_alpha_dw = t_alpha,
T1 = T1,
T2 = T2,
t_alpha_up = -t_alpha,
H01 = ifelse(T1 > t_alpha_up | T1 < t_alpha_dw, "Rejected", "Non-Rejected"),
H02 = ifelse(T2 > t_alpha_up | T2 < t_alpha_dw, "Rejected", "Non-Rejected")
) %>%
dplyr::mutate_if(is.numeric, format, digits = 4, scientific = FALSE)
colnames(kab) <- c("$$n$$", "$$\\alpha$$", "$$\\frac{N_n}{n}$$", "$$t_{\\alpha/2}$$",
"$$\\text{T}_1^{\\alpha}$$", "$$\\text{T}_2^{\\alpha}$$", "$$t_{\\alpha/2}$$", "$$H_0(\\text{T}_1)$$", "$$H_0(\\text{T}_2)$$")
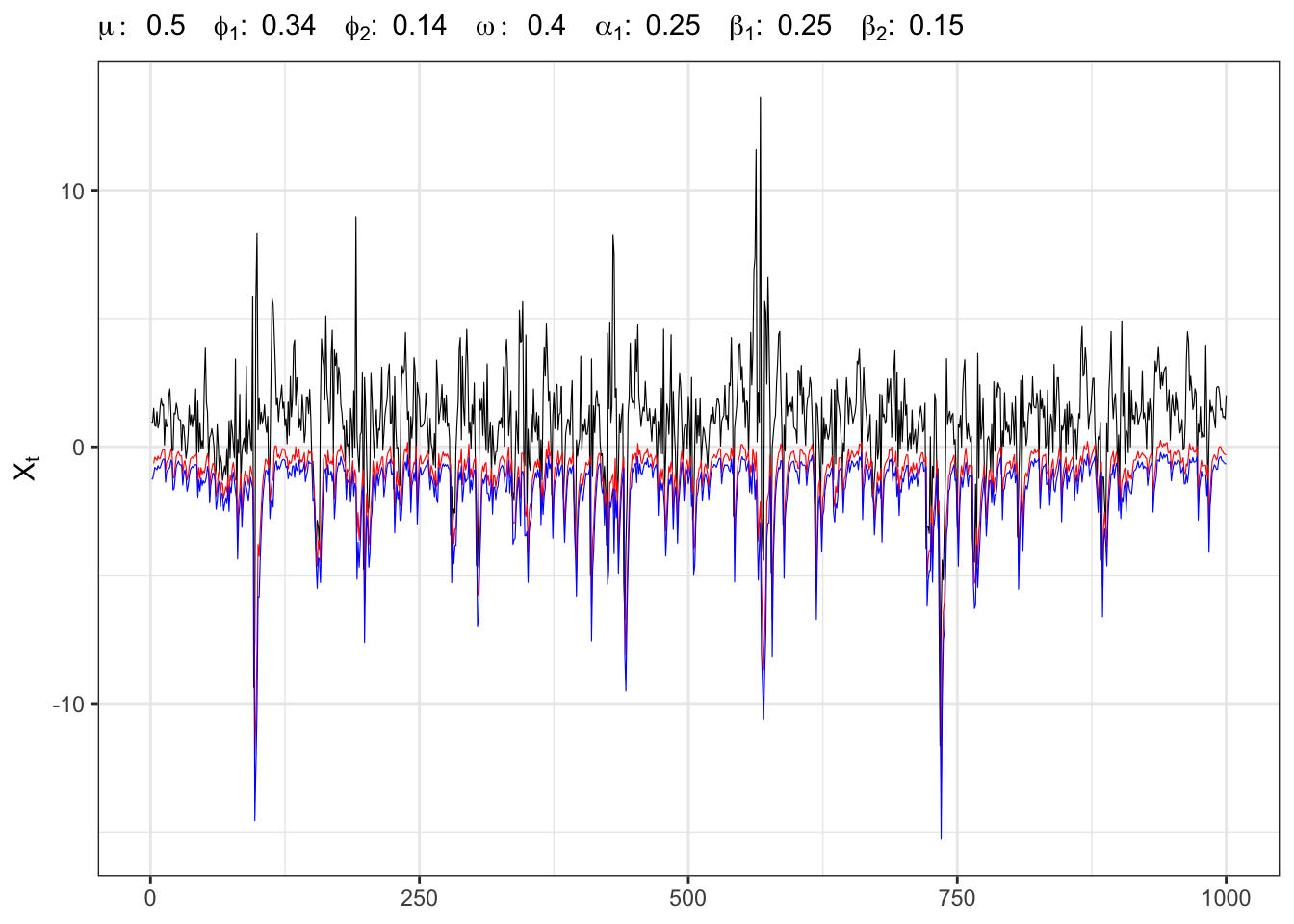
knitr::kable(kab)27.5 Example:
Instead of simulating exactly
AR(2)-GARCH(1,2) simulation and VaR
# ==================== Setups =====================
set.seed(1) # random seed
ci <- 0.05 # confidence level
t_bar <- 1000 # number of simulations
# parameters
parAR <- c(mu=0.5, phi1 = 0.34, phi1 = 0.14)
parGARCH <- c(omega=0.4, alpha1=0.25,
beta1=0.25, beta2=0.15)
# quasi long-term std deviation of residuals
sigma_eps <- sqrt(parGARCH[1]/(1-sum(parGARCH[-1])))
# ================== Simulation ===================
set.seed(1)
# Initial points
Xt <- rep(parAR[1]/(1-sum(parAR[-1])), 3)
mu <- rep(parAR[1]/(1-sum(parAR[-1])), 3)
sigma <- rep(sigma_eps, 3)
# Simulated residuals
eps <- rt(t_bar, 5)
eps[1:3] <- eps[1:3]*sigma_eps
# Value at Risk
q_alpha = qnorm(ci)
VaR = c(0)
for(t in 3:t_bar){
# AR component
mu[t] <- parAR[1] + parAR[2]*Xt[t-1] + parAR[3]*Xt[t-2]
# ARCH component
sigma[t] <- parGARCH[1] + parGARCH[2]*eps[t-1]^2
# GARCH component
sigma[t] <- sigma[t] + parGARCH[3]*sigma[t-1]^2 + parGARCH[4]*sigma[t-2]^2
sigma[t] <- sqrt(sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated value at risk
VaR[t] <- mu[t] + sigma[t]*q_alpha
# Simulated time series
Xt[t] <- mu[t] + eps[t]
}
# Empirical quantile
q_alpha_emp <- quantile(eps/sigma, probs = ci)
VaR_emp <- mu + sigma*q_alpha_emp
# ===================== Plot ======================
# GARCH(1,1) simulation
ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, VaR), size = 0.2, color = "red")+
geom_line(aes(1:t_bar, VaR_emp), size = 0.2, color = "blue")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", parAR[1],
"\\;\\; \\phi_{1}:\\;", parAR[2],
"\\;\\; \\phi_{2}:\\;", parAR[3],
"\\;\\; \\omega:\\;", parGARCH[1],
"\\;\\; \\alpha_{1}:\\;", parGARCH[2],
"\\;\\; \\beta_{1}:\\;", parGARCH[3],
"\\;\\; \\beta_{2}:\\;", parGARCH[4],
"$")))Computing the test on the normal quantile gives a rejection of the null hypothesis
VaR test Student-
# ======================================
# Violation of the VaR
vt <- ifelse(Xt < VaR, 1, 0)
vt[1:3] <- 0
# Theoric variance
v_theoric <- ci*(1-ci)
# Empiric variance
v_empiric <- (sum(vt)/t_bar)*(1 - sum(vt)/t_bar)
# Standardized number of violations
Nt <- sum((vt - ci)/sqrt(v_theoric))
# Statistic test (NV_1)
T1 <- (1/sqrt(t_bar))*Nt
# Statistic test (NV_2)
T2 <- (sqrt(v_theoric)/sqrt(v_empiric))*T1
# Rejection level
t_alpha <- qnorm(ci/2)
# =============== Kable ===============
kab <- dplyr::tibble(
n = t_bar,
alpha = paste0(format(ci*100, digits = 3), "%"),
alpha_hat = paste0(format(sum(vt)/t_bar*100, digits = 3), "%"),
t_alpha_dw = t_alpha,
T1 = T1,
T2 = T2,
t_alpha_up = -t_alpha,
H01 = ifelse(T1 > t_alpha_up | T1 < t_alpha_dw, "Rejected", "Non-Rejected"),
H02 = ifelse(T2 > t_alpha_up | T2 < t_alpha_dw, "Rejected", "Non-Rejected")
) %>%
dplyr::mutate_if(is.numeric, format, digits = 4, scientific = FALSE)
colnames(kab) <- c("$$n$$", "$$\\alpha$$", "$$\\frac{N_n}{n}$$", "$$-t_{\\alpha/2}$$",
"$$\\text{T}_1^{\\alpha}$$", "$$\\text{T}_2^{\\alpha}$$", "$$t_{\\alpha/2}$$",
"$$H_0(\\text{T}_1)$$", "$$H_0(\\text{T}_2)$$")
knitr::kable(kab)Setting